Decoding Lupus: How Genetic Clues Could Revolutionize Treatment
"New research reveals how copy number variations in our genes may hold the key to understanding and treating systemic lupus erythematosus."
Systemic lupus erythematosus (SLE), often simply called lupus, is a chronic autoimmune disease that can affect almost any part of the body. Characterized by inflammation and a wide range of symptoms, from fatigue and joint pain to skin rashes and organ damage, lupus can be challenging to diagnose and manage. While the exact causes of lupus remain elusive, it's widely accepted that genetics play a significant role in its development.
Recent advancements in genetic research are beginning to unravel the intricate web of factors contributing to lupus. A fascinating area of study is copy number variation (CNV), which refers to differences in the number of copies of specific genes within an individual's DNA. Unlike single-gene mutations, CNVs involve larger segments of DNA and can have profound effects on gene expression and function. New research is revealing how these CNVs may influence a person's susceptibility to lupus and potentially pave the way for more targeted therapies.
This article delves into the latest findings on CNVs and their connection to lupus. We will explore the groundbreaking study that examined the role of CNVs in a diverse population, the specific genes implicated, and the potential implications for future treatments. This information offers a fresh perspective on how genetics influences complex diseases like lupus and offers hope for a better future for those affected.
Unveiling the Genetic Puzzle: The Role of Copy Number Variations in Lupus

The research, published in PLOS ONE, focused on a specific type of genetic variation: copy number variations (CNVs). CNVs represent differences in the number of copies of a particular gene within a person's genome. This study employed a case-control design, comparing the genomes of individuals with lupus to those without the disease. The researchers used advanced genomic tools to detect and analyze CNVs across the entire genome.
- The study examined CNVs in SLE patients and healthy controls.
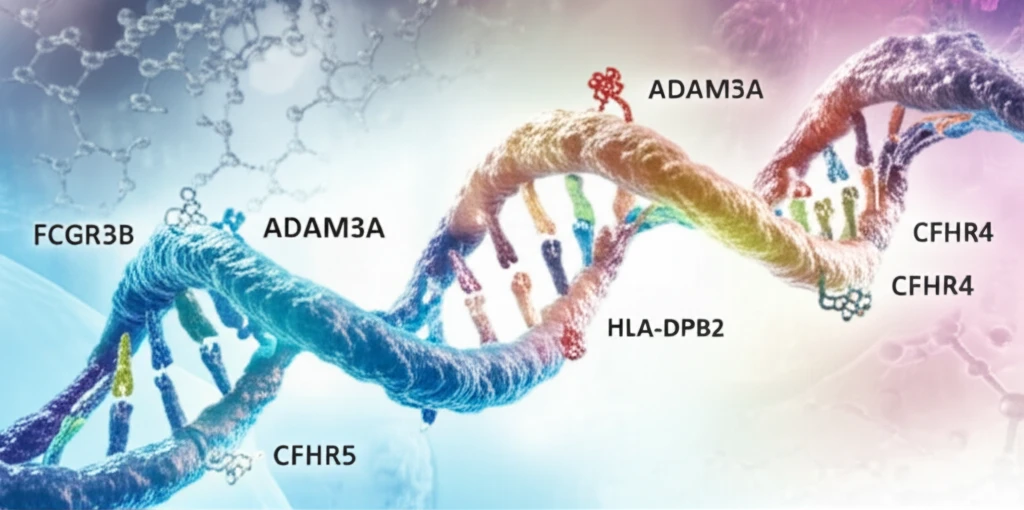
- Researchers found a synergistic effect between FCGR3B and ADAM3A genes.
- Deletions in both genes increased lupus risk.
- Rare CNVs were identified, including deletions in CFHR4, CFHR5, and HLA-DPB2.
A New Era for Lupus Treatment
The research discussed offers an exciting glimpse into the future of lupus treatment. By identifying specific genetic variations associated with the disease, scientists can develop more targeted therapies. For example, understanding the role of the FCGR3B and ADAM3A genes could lead to the development of drugs that specifically address the underlying genetic mechanisms contributing to lupus. Additionally, identifying individuals at higher genetic risk for lupus may enable earlier diagnosis and intervention, potentially improving outcomes and quality of life. This research is an important step towards personalized medicine for lupus and provides real hope for patients and their families.
